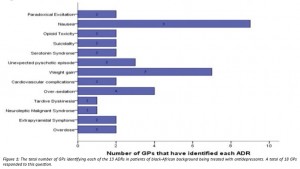
General practitioner awareness of pharmacogenomic testing and drug metabolism activity status amongst the Black-African population in the Greater Western Sydney region
Yashutosh Joshi, Abhishek Karavadra, Maiyoori Jeyaprakash
Tuesday, September 1st, 2015
Background: Individuals of black-African background have a high variability in drug metabolising enzyme polymorphisms. Consequently, unless these patients are tested for these polymorphisms, it becomes difficult to predict which patients may have a sub-therapeutic response to medications (such as anti- depressants) or experience an adverse drug reaction. Given the increasing population of black-Africans in Australia, GPs are on the front line of this issue, especially in Greater Western Sydney (GWS) – one of the country’s rapidly increasing populations due to migration. Aim: To ascertain the awareness of GPs regarding drug metabolising enzyme polymorphisms in the black-African population and pharmacogenomic testing in the GWS community. Methods: A descriptive, cross-sectional study was conducted in GWS by analysing GP responses to a questionnaire consisting of closed and open-ended questions. Results: A total of 46 GPs completed the questionnaire. It was found that 79.1% and 79.5% of respondents were unaware of: the high variability in drug metabolism enzyme activity in the black-African population and pharmacogenomic testing (respectively). No respondents had ever utilised pharmacogenomic testing. Only a small proportion of GPs “always” considered a patient’s genetic factors (13.9%) and enzyme metaboliser status (11.1%) in clinical practice. Preferred education media for further information included written material, direct information from other health professionals (such as pharmacists) and verbal teaching sessions. Conclusion: There was a low level of awareness of enzyme metaboliser status and pharmacogenomic testing amongst GPs in GWS. A future recommendation to ameliorate this includes further education provision through a variety of media noted in the study.

Introduction
Depression accounts for 13% of Australia’s total disease burden, making it an important health issue in the current context. [1] General Practitioners (GPs) are usually the first point of contact for patients seeking help for depression. [2,3] Antidepressant prescription is the most common treatment form for depression in Australia with GPs prescribing an antidepressant to treat up to 40% of all psychological problems. [2] This makes GP awareness of possible treatment resistance or adverse drug reactions (ADRs) to these medications vital.
Binder et al. [4] described pharmacogenomics as “the use of genome- wide approaches to elucidate individual differences in the outcome of drug therapy”. Detecting clinically relevant polymorphisms in genetic expression can potentially be used to identify susceptibility to ADRs. [4] This would foster the application of personalised medicine by encouraging an inter-individual approach to medication and dose prescriptions based on an individual’s predicted response to medications. [4,5]
Human DNA contains genes that code for 57 cytochrome (CYP) P450 isoenzymes; these are a clinically important family of hepatic and gastrointestinal isoenzymes responsible for the metabolism of over 70% of clinically prescribed drugs. [5-10] The CYP family of enzymes are susceptible to polymorphisms as a result of genetic variations, influenced by factors such as ethnicity. [6,5,10] Research has shown that polymorphisms in certain CYP drug metabolising enzymes can result in phenotypes that class individuals as “ultrarapid metabolisers (UMs), extensive metabolisers (EMs), intermediate metabolisers (IMs) and poor metabolisers (PMs).”[6,10] These categories are clinically important as they determine whether or not a drug stays within the therapeutic range. Individuals with PM status may be susceptible to experiencing ADRs as a result of toxicity, and conversely, those with UM status may not receive a therapeutic effect. [5,6,10,11]
When considering the metabolism of antidepressants, the highly polymorphic CYP enzymes: CYP2C19 and CYP2D6 are known to be involved. [5,10,12] A study by Xie et al. [13] has shown that for the CYP2D6 enzyme alone, allelic variations induce polymorphisms that result in a PM phenotype of “~1%” in Asian populations, “0-5%” among Caucasians and a variation of between “0-19%” in black- African populations. This large disparity of polymorphism phenotypes was reproduced in a recent study, which also showed that the variation is not exclusive to the CYP2D6 enzyme. [6] It has been reported that the incidence of ADRs among PMs treated with drugs such as antidepressants is 44% compared to 21% in other patients. [5,14] Consequently, increased costs have been associated with the management of UM or PM patients. [5]
The black-African population in Australia and specifically Sydney (where GWS is one of the fastest growing regions) continues to rise through migration and humanitarian programs. [15-18] Almost 30% of Africans settling in Australia in the decade leading to the year 2007 did so under humanitarian programs including under refugee status. [15-17] As refugees are at a higher risk of having mental health problems including depression due to their traumatic histories and post-migratory difficulties, GPs in GWS face increased clinical interactions with black-Africans at risk of depression. [19,20] Considering the high variability of enzyme polymorphisms in this population, pharmacogenomic testing may play a role in the primary care of these patients. We therefore conducted a study to assess GP awareness of pharmacogenomic testing and the differences in enzyme metaboliser status (drug metabolism phenotypes). We also investigated the GP preferences of media for future education on these topics.
Methodology
Study Design and Setting
This is a descriptive, cross-sectional study. Ethics approval was granted by the Human Research Ethics Committee.
Considering GWS is the fastest growing region in Sydney, we focussed on particular suburbs in GWS (Blacktown, Parramatta and Holroyd Local Government Areas). [17-20] Using geographical cluster sampling, a list of GP practices were identified with the aim of recruiting 50 participants.
Study tool
Data was collected using a questionnaire validated by university supervisors and designed to elicit the level of understanding and awareness among GPs. The main themes of the questionnaire involved: questions regarding basic demographic information; questions aimed at determining the level of GP awareness regarding differences in drug metabolising phenotypes and pharmacogenomic testing; and open- ended questions eliciting the preferred methods of education with respect to pharmacogenomic testing.
Data Collection
We invited 194 GPs between April and May 2014 to participate in the study. The questionnaire and participant information sheet were either given to the practice managers or to the GPs in person. Questionnaires were collected in person within the following two weeks.
Data Analysis
Data was analysed using SPSS (version 22, IBM Australia). Descriptive statistics were used to summarise findings, with p-values calculated using Chi-square analysis (with Yates correction) to compare two sets of data. A p-value of <0.05 indicated statistical significance.
Results
The overall response rate was 23.7% (46/194). Our respondents included: 27 females and 19 males. The mean number of years of experience in general practice was 13.9 and most GPs (93.4%, 43/46) had received some form of training in antidepressant prescription in the last 5 years. The number of patients of black-African background seen in the last 6 months ranged from 0 to greater than 100. Only
26.1% (12/46) of GPs reported no consultations with a patient of black- African background within this timeframe. Of the 73.9% (34/46) of GPs who had seen at least one patient from this cohort, 55.9% (19/34) had treated at least one patient for depression with antidepressants.
GPs experience of ADRs in patients of black-African background treated for depression
From 46 participants, 19 had treated a patient of black-African background with antidepressants, 18/19 reported having identified at least one ADR (Figure 1).
GP awareness and consideration of drug metabolism activity status and genetic factors
Awareness amongst GPs of the different drug metabolism activity phenotypes in black-Africans was low with 79.1% (34/43) being unaware. Patients’ genetic factors and enzyme metaboliser status were “always” considered by only 13.9% (5/36) and 11.1% (4/36) of GPs, respectively. There was no statistically significant difference regarding awareness between GPs who had treated black-African patients and those who had not (21.1% vs 13.3% respectively, p=0.89).
GP awareness and use of pharmacogenomic testing
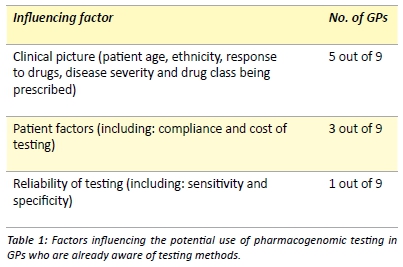
The awareness of methods for testing a patient’s key drug metabolising enzymes, also known as pharmacogenomic testing, was extremely low with 79.5% (35/44) of GPs being unaware of the testing methods available in Australia. Of the 20.5% of GPs (9/44) who were aware, none had utilised pharmacogenomic testing for their black-African patients. These nine GPs then nominated factors that would influence their utilisation of pharmacogenomic testing on these individuals. Three main categories of influence emerged (Table 1). When specifically asked whether they would be more inclined to utilise pharmacogenomic testing on black-African patients who had previously experienced ADRs, 88.9% (8/9) GPs stated that they would be more inclined.
Preferred education media
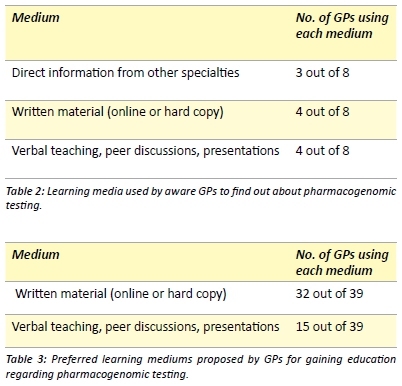
GPs that were aware of pharmacogenomic testing were asked, through an open-ended question, how they obtained information regarding these methods. Three main categories were identified based on their responses (Table 2). All GPs were then asked to note down their preferred medium of education for pharmacogenomic testing (Table 3). Multiple responses were allowed.
Discussion
This study showed that there is a low level of awareness regarding pharmacogenomic testing and the differences in drug metabolism phenotypes among GPs. Additionally, we identified the preferred education media for providing information to GPs (Table 3). Awareness of pharmacogenomic testing and of the differences in drug enzyme metaboliser status (phenotype) could be valuable in the clinical setting. Improved patient outcomes have been noted when doctors are able to personalise management based on information from pharmacogenomic testing,[21] with Hall-Flavin et al. [21] noting significantly improved baseline depression scores amongst patients with depression whose doctors were provided with information on pharmacogenomics.
A previous study reported that a high proportion (97.6%) of physicians agreed that differences in genetic factors play a major role in drug responses. [22] Whilst it is arguable that knowledge of genetic factors holistically playing a role in drug response may be universal, our study specifically focussed on the knowledge of differences in enzyme metaboliser status. It was found that 79.1% of GPs (34/43) were unaware, with only a small number of GPs “always” considering enzyme metaboliser status (11.1%) in their management. Given the aforementioned importance of genetic factors and the potential to reduce ADRs using personalised medicine, this is an area for improvement.
When considering pharmacogenomic testing, we found 79.5% (35/44) of GPs to be unaware of testing methods. No GP had ever utilised pharmacogenomic testing, this low rate of utilisation is also reported previously in other several studies. [22-24] A lack of utilisation and awareness arguably forms a barrier against the effective incorporation of personalised medicine in the primary care setting. These low figures represent a lack of education regarding pharmacogenomics and its clinical applications. This is an issue that has been recognised since the arrival of these testing methods. [25] McKinnon et al. [25] highlighted that this lack of education across healthcare professionals is significant enough to be considered a “barrier to the widespread uptake of pharmacogenomics”. To ameliorate the situation, the International Society of Pharmacogenomics has issued recommendations in 2005 for pharmacogenomics to be incorporated into medical curricula. [26] Another contributing factor to the low utilisation of testing could include the lack of subsidised tests available through Medicare. Currently, pathology labs do provide pharmacogenomic testing (such as Douglas Hanley Moir and Healthscope), however this is largely done so through the patient’s expenses as only two methods are subsidised by Medicare. [23,27,28]
Amongst those aware of pharmacogenomic testing, eight out of nine GPs answered that they would be more likely to utilise pharmacogenomic testing in black-African patients who had previously experienced ADRs; this is consistent with findings noted by van Puijenbroek et al. [29]. Among these GPs, factors that were noted to be potential influences in their utilisation of testing included: patient factors such as compliance and the reliability of the test, and, factors affecting the clinical picture (as described in Table 1). This is consistent with findings by studies that have also identified cost and a patient’s individual response to drugs as influential factors in a physician’s decision making. [29,30]
Considering that the majority of information regarding enzyme metabolism and pharmacogenomic testing was published in pharmacological journals,[6,8-14,30-32] much of this knowledge may not have been passed on to GPs. In order to understand the preferred media of information for GPs, we posed open-ended questions and discovered that the majority of GPs who answered the question (32/39), would prefer information in the form of writing (Table 3). This could be either in the form of online sources (such as guidelines, summaries, the National Prescribing Service or the Monthly Index of Medical Specialities) or peer reviewed journal articles. Current literature also reflects this preference for GPs to gain education regarding pharmacogenomics through journal articles. [22] The other preferred medium of education was through verbal teachings, peer discussions and presentations (Table 3), with there being specific interest in information being disseminated by clinical pathology laboratories; this is also reflected in the literature. [22,29]
Strengths and limitations
Small sample size is a limitation of this study with possible contributing factors including: the short amount of time allowed for data collection and the low response rate due to GP time constraints. Strengths of the study include the use of a validated questionnaire catered to our target population and open-ended questions which gave us further insight into GP preferences.
Implications and future research
Currently, anti-coagulants provide an example of the clinical applications of considering enzyme polymorphisms in patient management. [33,34] Warfarin is a particular example where variability in INR has been associated with enzyme polymorphisms, leading to the utilisation of dosage algorithms to optimise clinical outcomes. [34] Similarly, when using antidepressants, pharmacogenomic testing could play a role in clinical decision making with Samer et al. [5] suggesting dose reductions and serum monitoring for those with known PM status. However, as identified in our study, there is an overall lack of awareness regarding the differences in enzyme metaboliser status and the methods available for pharmacogenomic testing.
Future studies should focus on the clinical practicality of utilising these tests. Additionally, future studies should determine the effectiveness of the identified GP preferred modalities of education in raising awareness.
Conclusion
There is a low awareness among GPs regarding both the differences in enzyme metaboliser status in the black-African community, and the methods of pharmacogenomic testing.
To optimise clinical outcomes in black-African patients with depression, it may be useful to inform GPs of the availability and application of pharmacogenomic testing. We have highlighted the preferred education modalities through which this may be possible.
Acknowledgements
We would like to acknowledge and thank Dr. Irina Piatkov for her support as a supervisor during this project.
Conflict of interest
None declared.
Correspondence
Y Joshi: 17239266@student.uws.edu.au
References
[1] Australian Institute of Health and Welfare. The burden of disease and injury in Australia 2003 [Internet]. 2007 [cited 2014 April 25]. Available from: http://www.aihw.gov.au/ publication-detail/?id=6442467990
[2] Charles J, Britt H, Fahridin S, Miller G. Mental health in general practice. Aust Fam Physician. 2007;36(3):200-1.
[3] Pierce D, Gunn J. Depression in general practice: consultation duration and problem solving therapy. Aust Fam Physician. 2011;40(5):334-6.
[4] Binder EB, Holsboer F. Pharmacogenomics and antidepressant drugs. Ann Med. 2006;38(2):82-94.
[5] Samer CF, Lorenzini KI, Rollason V, Daali Y, Desmeules JA. Applications of CYP450 testing in the clinical setting. Mol Diagn Ther. 2013;17(3):165-84.
[6] Alessandrini M, Asfaha S, Dodgen MT, Warnich L, Pepper MS. Cytochrome P450 pharmacogenetics in African populations. Drug Metab Rev. 2013;45(2):253-7.
[7] Yang X, Zhang B, Molony C, Chudin E, Hao K, Zhu J et al. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010;20(8):1020-36.
[8] Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities and impact of genetic variation. Pharmacol Therapeut. 2013;138(1):103-41.
[9] Guengerich FP. Cytochrome P450 and chemical toxicology. Chem Res Toxicol. 2008;21(1):70-83.
[10] Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6-13.
[11] Zhou S. Polymorphism of human cytochrome P450 2D6 and its clinical significance. Clin Pharmacokinet. 2009;48(11):689-723.
[12] Li-Wan-Po A, Girard T, Farndon P, Cooley C, Lithgow J. Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br J Clin Pharmacol. 2010;69(3):222-30.
[13] Xie HG, Kim RB, Wood AJJ, Stein CM. Molecular Basis of ethnic differences in drug disposition and response. Ann Rev Pharmacol Toxicol. 2001;41:815-50.
[14] Chen S, Chou WH, Blouin RA, Mao Z, Humphries LL, Meek QC et al. The cytochrome P450 2D6 (CYP2D6) enzyme polymorphism: screening costs and influence on clinical outcomes in psychiatry. Clin Pharmacol Ther. 1996;60(5):522–34.
[15] Hugo G. Migration between Africa and Australia: a demographic perspective – Background paper for African Australians: A review of human rights and social inclusion issues. Australian Human Rights Commission [Internet]. 2009 Dec [cited 2014 April 26]. Available from: https://www.humanrights.gov.au/sites/default/files/content/Africanaus/papers/Africanaus_paper_hugo.pdf
[16] Joint Standing Committee on Foreign Affairs, Defence and Trade. Inquiry into Australia’s relationship with the countries of Africa [Internet]. 2011 [cited 2014 April 26]. Available from: http://www.aph.gov.au/Parliamentary_Business/Committees/House_of_Representatives_Committees?url=jfadt/africa%2009/report.htm
[17] Census 2006 – People born in Africa [Internet]. Australian Bureau of Statistics; 2008 August 20 [updated 2009 April 14; cited 2014 April 26]. Available from: http://www.abs.gov.au/AUSSTATS/abs@.nsf/Lookup/3416.0Main+Features32008
[18] Greater Western Sydney Economic Development Board. Some national transport and freight infrastructure priorities for Greater Western Sydney [Internet]. Infrastructure Australia; 2008 [cited April 25 2014]. Available from: http:// w w w. i n fras tru ctu r eau s tral i a. g o v. au /p u b l i c_su b mi ssi o ns/p u b l i sh ed /fi l es/368_ greaterwesternsydneyeconomicdevelopmentboard_SUB.pdf
[19] Furler J, Kokanovic R, Dowrick C, Newton D, Gunn J, May C. Managing depression among ethnic communities: a qualitative study. Ann Fam Med. 2010;8:231-6.
[20] Robjant K, Hassan R, Katona C. Mental health implications of detaining asylum seekers: systematic review. Br J Psychiatry. 2009;194:306-12.
[21] Hall-Flavin DK, Winner JG, Allen JD, Carhart JM, Proctor B, Snyder KA et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2013;23(10):535- 48.
[22] Stanek EJ, Sanders CL, Taber KA, Khalid M, Patel A, Verbrugge RR et al. Adoption of pharmacogenomics testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91(3):450-8.
[23] Sheffield LJ, Phillimore HE. Clinical use of pharmacogenomics tests in 2009. Clin Biochem Rev. 2009;30(2):55-65.
[24] Corkindale D, Ward H, McKinnon R. Low adoption of pharmacogenetic testing: an exploration and explanation of the reasons in Australia. Pers Med. 2007;4(2):191-9.
[25] McKinnon R, Ward M, Sorich M. A critical analysis of barriers to the clinical implementation of pharmacogenomics. Ther Clin Risk Manag. 2007;3(5):751-9.
[26] Gurwitz D, Lunshof J, Dedoussis G, Flordellis C, Fuhr U, Kirchheiner J et al. Pharmacogenomics education: International Society of Pharmacogenomics recommendations for medical, pharmaceutical, and health schools deans of education. Pharmacogenomics J. 2005;5(4):221-5.
[27] Pharmacogenomics [Internet]. Healthscope Pathology; 2014 [cited 2014 October 22] Available from: http://www.healthscopepathology.com.au/index.php/advanced pathology/pharmacogenomics/
[28] Overview of Pharmacogenomic testing. Douglas Hanley Moir Pathology; 2013 [cited 2014 October 22]. Available from: http://www.dhm.com.au/media/21900626/pharmacogenomics_brochure_2013_web.pdf
[29] van Puijenbroek E, Conemans J, van Groostheest K. Spontaneous ADR reports as a trigger for pharmacogenetic research: a prospective observational study in the Netherlands. Drug Saf. 2009;32(3):225-64.
[30] Rogausch A, Prause D, Schallenberg A, Brockmoller J, Himmel W. Patients’ and physicians’ perspectives on pharmacogenetic testing. Pharmacogenomics. 2006;7(1):49- 59.
[31] Akilillu E, Persson I, Bertilsson L, Johansson I, Rodrigues F, Ingelman-Sundberg M. Frequent distribution of ultrarapid metabolizers of debrisoquine in Ethopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278(1):441-6.
[32] Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229-43.
[33] Cresci S, Depta JP, Lenzini PA, Li Ay, Lanfear DE, Province MA et al. Cytochrome p450 gene variants, race, and mortality among clopidogrel-treated patients after acute myocardial infarction. Circ Cardiovasc Genet. 2014 7(3):277-86.
[34] Becquemont L. Evidence for a pharmacogenetic adapted dose of oral anticoagulant in routine medical practice. Eur J Clin Pharmacol. 2008 64(10):953-60